The Potential of Cell-Penetrating Peptides for mRNA Delivery to Cancer Cells
Abstract
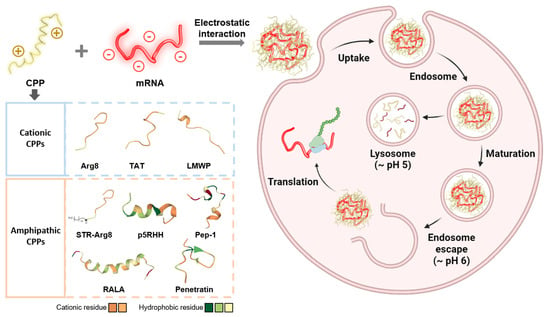
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. CPP/mRNA Complex Formation and Characterization
2.3. Cell Cultures
2.4. In Vitro Cytotoxicity of CPP/mRNA Complexes
2.5. Cellular Uptake of CPP/mRNA Complexes
2.6. Evaluation of CPP/mRNA-Mediated Protein Expression
2.7. Statistical Analysis
3. Results
3.1. Optimization of CPP/mRNA Complex Formation
3.2. Screening of CPPs Suitable for Complexation with mRNA
3.3. Intracellular Cytotoxicity of CPP/mRNA Complexes
3.4. In Vitro Cellular Uptake of CPP/mRNA Complexes
3.5. Evaluation of CPP/mRNA-Mediated Protein Expression
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shahbazi, R.; Ozpolat, B.; Ulubayram, K. Oligonucleotide-based theranostic nanoparticles in cancer therapy. Nanomedicine 2016, 11, 1287–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginn, S.L.; Amaya, A.K.; Alexander, I.E.; Edelstein, M.; Abedi, M.R. Gene therapy clinical trials worldwide to 2017: An update. J. Gene Med. 2018, 20, e3015. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Shi, Q.; Zhang, H.; Yang, K.; Ke, Y.; Wang, Y.; Qiao, L. Advances in the techniques and methodologies of cancer gene therapy. Discov. Med. 2019, 27, 45–55. [Google Scholar] [PubMed]
- Yamada, Y. Nucleic Acid Drugs-Current Status, Issues, and Expectations for Exosomes. Cancers 2021, 13, 5002. [Google Scholar] [CrossRef]
- Kramps, T.; Elbers, K. Introduction to RNA Vaccines. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; Volume 1499, pp. 1–11. [Google Scholar] [CrossRef]
- Cavazzana, M.; Six, E.; Lagresle-Peyrou, C.; Andre-Schmutz, I.; Hacein-Bey-Abina, S. Gene Therapy for X-Linked Severe Combined Immunodeficiency: Where Do We Stand? Hum. Gene Ther. 2016, 27, 108–116. [Google Scholar] [CrossRef] [Green Version]
- Iavarone, C.; O’Hagan, D.T.; Yu, D.; Delahaye, N.F.; Ulmer, J.B. Mechanism of action of mRNA-based vaccines. Exp. Rev. Vaccines 2017, 16, 871–881. [Google Scholar] [CrossRef]
- Wadhwa, A.; Aljabbari, A.; Lokras, A.; Foged, C.; Thakur, A. Opportunities and Challenges in the Delivery of mRNA-based Vaccines. Pharmaceutics 2020, 12, 102. [Google Scholar] [CrossRef] [Green Version]
- Miao, L.; Zhang, Y.; Huang, L. mRNA vaccine for cancer immunotherapy. Mol. Cancer 2021, 20, 41. [Google Scholar] [CrossRef]
- Batich, K.A.; Mitchell, D.A.; Healy, P.; Herndon, J.E., 2nd; Sampson, J.H. Once, Twice, Three Times a Finding: Reproducibility of Dendritic Cell Vaccine Trials Targeting Cytomegalovirus in Glioblastoma. Clin. Cancer Res. 2020, 26, 5297–5303. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [Green Version]
- Let’s talk about lipid nanoparticles. Nat. Rev. Mater. 2021, 6, 99. [CrossRef]
- Kowalski, P.S.; Rudra, A.; Miao, L.; Anderson, D.G. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther. 2019, 27, 710–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibba, M.L.; Ciccone, G.; Esposito, C.L.; Catuogno, S.; Giangrande, P.H. Advances in mRNA non-viral delivery approaches. Adv. Drug Deliv. Rev. 2021, 177, 113930. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Kariko, K.; Tureci, O. mRNA-based therapeutics—Developing a new class of drugs. Nat. Rev. Drug Discov. 2014, 13, 759–780. [Google Scholar] [CrossRef]
- Niculescu, A.G.; Birca, A.C.; Grumezescu, A.M. New Applications of Lipid and Polymer-Based Nanoparticles for Nucleic Acids Delivery. Pharmaceutics 2021, 13, 2053. [Google Scholar] [CrossRef]
- Hajj, K.A.; Whitehead, K.A. Tools for translation: Non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017, 2, 17056. [Google Scholar] [CrossRef]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef]
- Meng, C.; Chen, Z.; Li, G.; Welte, T.; Shen, H. Nanoplatforms for mRNA Therapeutics. Adv. Ther. 2021, 4, 2000099. [Google Scholar] [CrossRef]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef]
- Hassett, K.J.; Higgins, J.; Woods, A.; Levy, B.; Xia, Y.; Hsiao, C.J.; Acosta, E.; Almarsson, O.; Moore, M.J.; Brito, L.A. Impact of lipid nanoparticle size on mRNA vaccine immunogenicity. J. Control Release 2021, 335, 237–246. [Google Scholar] [CrossRef]
- Gaviria, M.; Kilic, B. A network analysis of COVID-19 mRNA vaccine patents. Nat. Biotechnol. 2021, 39, 546–548. [Google Scholar] [CrossRef] [PubMed]
- Boisguerin, P.; Konate, K.; Josse, E.; Vives, E.; Deshayes, S. Peptide-Based Nanoparticles for Therapeutic Nucleic Acid Delivery. Biomedicines 2021, 9, 583. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.P.; Arami, H.; Banga, I.; Gupta, J.; Gandhi, S. Cell penetrating peptides in preclinical and clinical cancer diagnosis and therapy. Oncotarget 2018, 9, 37252–37267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoo, H.; Oba, M.; Uchida, S. Cell-Penetrating Peptides: Emerging Tools for mRNA Delivery. Pharmaceutics 2021, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- Alves, I.D.; Goasdoue, N.; Correia, I.; Aubry, S.; Galanth, C.; Sagan, S.; Lavielle, S.; Chassaing, G. Membrane interaction and perturbation mechanisms induced by two cationic cell penetrating peptides with distinct charge distribution. Biochim. Biophys. Acta 2008, 1780, 948–959. [Google Scholar] [CrossRef]
- Varkouhi, A.K.; Scholte, M.; Storm, G.; Haisma, H.J. Endosomal escape pathways for delivery of biologicals. J. Control Release 2011, 151, 220–228. [Google Scholar] [CrossRef]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef]
- Madani, F.; Lindberg, S.; Langel, U.; Futaki, S.; Graslund, A. Mechanisms of cellular uptake of cell-penetrating peptides. J. Biophys. 2011, 2011, 414729. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, S.; Nakase, I.; Kawabata, N.; Yu, H.H.; Pujals, S.; Imanishi, M.; Giralt, E.; Futaki, S. Cytosolic targeting of macromolecules using a pH-dependent fusogenic peptide in combination with cationic liposomes. Bioconjug. Chem. 2009, 20, 953–959. [Google Scholar] [CrossRef]
- van den Brand, D.; Gorris, M.A.J.; van Asbeck, A.H.; Palmen, E.; Ebisch, I.; Dolstra, H.; Hallbrink, M.; Massuger, L.; Brock, R. Peptide-mediated delivery of therapeutic mRNA in ovarian cancer. Eur. J. Pharm. Biopharm. 2019, 141, 180–190. [Google Scholar] [CrossRef]
- Agrawal, P.; Bhalla, S.; Usmani, S.S.; Singh, S.; Chaudhary, K.; Raghava, G.P.; Gautam, A. CPPsite 2.0: A repository of experimentally validated cell-penetrating peptides. Nucleic Acids Res. 2016, 44, D1098–D1103. [Google Scholar] [CrossRef] [PubMed]
- Ignatovich, I.A.; Dizhe, E.B.; Pavlotskaya, A.V.; Akifiev, B.N.; Burov, S.V.; Orlov, S.V.; Perevozchikov, A.P. Complexes of plasmid DNA with basic domain 47-57 of the HIV-1 Tat protein are transferred to mammalian cells by endocytosis-mediated pathways. J. Biol. Chem. 2003, 278, 42625–42636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Won, Y.W.; Adhikary, P.P.; Lim, K.S.; Kim, H.J.; Kim, J.K.; Kim, Y.H. Oligopeptide complex for targeted non-viral gene delivery to adipocytes. Nat. Mater. 2014, 13, 1157–1164. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Pan, D.; Qiu, F.; Sun, P.; Zhang, Y.H. Selective DNA delivery to tumor cells using an oligoarginine-LTVSPWY peptide. PLoS ONE 2014, 9, e110632. [Google Scholar] [CrossRef]
- Choi, Y.S.; Lee, J.Y.; Suh, J.S.; Kwon, Y.M.; Lee, S.J.; Chung, J.K.; Lee, D.S.; Yang, V.C.; Chung, C.P.; Park, Y.J. The systemic delivery of siRNAs by a cell penetrating peptide, low molecular weight protamine. Biomaterials 2010, 31, 1429–1443. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Ye, J.; Pei, X.; Sun, L.; Liu, E.; Wang, J.; Huang, Y.; Lee, S.J.; He, H. Improved method for synthesis of low molecular weight protamine-siRNA conjugate. Acta Pharm. Sin. B 2018, 8, 116–126. [Google Scholar] [CrossRef]
- Suh, J.S.; Lee, J.Y.; Choi, Y.S.; Chung, C.P.; Park, Y.J. Peptide-mediated intracellular delivery of miRNA-29b for osteogenic stem cell differentiation. Biomaterials 2013, 34, 4347–4359. [Google Scholar] [CrossRef]
- Udhayakumar, V.K.; De Beuckelaer, A.; McCaffrey, J.; McCrudden, C.M.; Kirschman, J.L.; Vanover, D.; Van Hoecke, L.; Roose, K.; Deswarte, K.; De Geest, B.G.; et al. Arginine-Rich Peptide-Based mRNA Nanocomplexes Efficiently Instigate Cytotoxic T Cell Immunity Dependent on the Amphipathic Organization of the Peptide. Adv. Healthc. Mater 2017, 6, 1601412. [Google Scholar] [CrossRef]
- Lockhart, J.H.; VanWye, J.; Banerjee, R.; Wickline, S.A.; Pan, H.; Totary-Jain, H. Self-assembled miRNA-switch nanoparticles target denuded regions and prevent restenosis. Mol. Ther. 2021, 29, 1744–1757. [Google Scholar] [CrossRef]
- Guo, Z.; Peng, H.; Kang, J.; Sun, D. Cell-penetrating peptides: Possible transduction mechanisms and therapeutic applications. Biomed. Rep. 2016, 4, 528–534. [Google Scholar] [CrossRef] [Green Version]
- Rydberg, H.A.; Matson, M.; Amand, H.L.; Esbjorner, E.K.; Norden, B. Effects of tryptophan content and backbone spacing on the uptake efficiency of cell-penetrating peptides. Biochemistry 2012, 51, 5531–5539. [Google Scholar] [CrossRef] [PubMed]
- Wadia, J.S.; Dowdy, S.F. Protein transduction technology. Curr. Opin. Biotechnol. 2002, 13, 52–56. [Google Scholar] [CrossRef]
- Konate, K.; Lindberg, M.F.; Vaissiere, A.; Jourdan, C.; Aldrian, G.; Margeat, E.; Deshayes, S.; Boisguerin, P. Optimisation of vectorisation property: A comparative study for a secondary amphipathic peptide. Int. J. Pharm. 2016, 509, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Konate, K.; Dussot, M.; Aldrian, G.; Vaissiere, A.; Viguier, V.; Neira, I.F.; Couillaud, F.; Vives, E.; Boisguerin, P.; Deshayes, S. Peptide-Based Nanoparticles to Rapidly and Efficiently “Wrap ‘n Roll” siRNA into Cells. Bioconjug. Chem. 2019, 30, 592–603. [Google Scholar] [CrossRef]
- Deshayes, S.; Decaffmeyer, M.; Brasseur, R.; Thomas, A. Structural polymorphism of two CPP: An important parameter of activity. Biochim. Biophys. Acta—Biomembr. 2008, 1778, 1197–1205. [Google Scholar] [CrossRef] [Green Version]
- Kalafatovic, D.; Giralt, E. Cell-Penetrating Peptides: Design Strategies beyond Primary Structure and Amphipathicity. Molecules 2017, 22, 1929. [Google Scholar] [CrossRef] [Green Version]
- Mo, R.H.; Zaro, J.L.; Shen, W.C. Comparison of cationic and amphipathic cell penetrating peptides for siRNA delivery and efficacy. Mol. Pharm. 2012, 9, 299–309. [Google Scholar] [CrossRef]
- Walrant, A.; Bauza, A.; Girardet, C.; Alves, I.D.; Lecomte, S.; Illien, F.; Cardon, S.; Chaianantakul, N.; Pallerla, M.; Burlina, F.; et al. Ionpair-pi interactions favor cell penetration of arginine/tryptophan-rich cell-penetrating peptides. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183098. [Google Scholar] [CrossRef]
- Fernando, D.; Sulthana, S.; Vasquez, Y. Cellular Uptake and Cytotoxicity of Varying Aspect Ratios of Gold Nanorods in HeLa Cells. ACS Appl. Bio Mater. 2020, 3, 1374–1384. [Google Scholar] [CrossRef]
- Hou, K.K.; Pan, H.; Ratner, L.; Schlesinger, P.H.; Wickline, S.A. Mechanisms of nanoparticle-mediated siRNA transfection by melittin-derived peptides. ACS Nano 2013, 7, 8605–8615. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.W.; Lee, H.J.; Tolliver, L.M.; Aronstam, R.S. Delivery of nucleic acids and nanomaterials by cell-penetrating peptides: Opportunities and challenges. Biomed. Res. Int. 2015, 2015, 834079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, M.; Huang, Y.W.; Aronstam, R.S.; Lee, H.J. Cellular delivery of noncovalently-associated macromolecules by cell-penetrating peptides. Curr. Pharm. Biotechnol. 2014, 15, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Bohmova, E.; Machova, D.; Pechar, M.; Pola, R.; Venclikova, K.; Janouskova, O.; Etrych, T. Cell-penetrating peptides: A useful tool for the delivery of various cargoes into cells. Physiol. Res. 2018, 67, S267–S279. [Google Scholar] [CrossRef] [PubMed]






| CPP | MW (Da) | Net Charge at pH 7.0 | Amino Acid Sequence | |
|---|---|---|---|---|
| Cationic CPPs | Arg8 | 1267.0 | +8 | RRRRRRRR |
| TAT | 1621.9 | +8 | GRKKRRQRRRPQ | |
| LMWP | 1879.1 | +10 | VSRRRRRRGGRRRR | |
| Amphipathic CPPs | STR-Arg8 | 1533.0 | +8 | Stearyl-RRRRRRRR |
| p5RHH | 2540.4 | +5 | VLTTGLPALISWIRRRHRRHC | |
| RALA | 3325.9 | +5 | WEARLARALARALARHLARALARALRACEA | |
| Pep-1 | 2848.2 | +3 | KETWWETWWTEWSQPKKKRKV | |
| Penetratin | 2248.2 | +7 | RQIKIWFQNRRMKWKK |
| mRNA/CPP | N/P Ratio | Particle Size (nm) | PDI | Zeta Potential |
|---|---|---|---|---|
| Arg8 | 1.5 | 158.3 (±0.0) | 0.09 | +16.9 (±0.2) |
| TAT | 3.9 | 141.7 (±0.9) | 0.07 | +16.5 (±0.9) |
| LMWP | 1.9 | 129.5 (±0.3) | 0.22 | +22.5 (±0.0) |
| STR-Arg8 | 7.8 | 156.8 (±4.7) | 0.28 | +28.6 (±0.3) |
| p5RHH | 9.7 | 154.3 (±0.1) | 0.20 | +16.8 (±0.9) |
| RALA | 9.7 | 174.6 (±3.7) | 0.23 | +20.2 (±1.7) |
| Pep-1 | 11.6 | 644.0 (±5.4) | 0.40 | +7.7 (±0.2) |
| Penetratin | 13.6 | 487.0 (±5.3) | 0.29 | +15.7 (±1.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.; Kim, H.; Kim, E.H.; Jang, H.; Jang, Y.; Chi, S.-G.; Yang, Y.; Kim, S.H. The Potential of Cell-Penetrating Peptides for mRNA Delivery to Cancer Cells. Pharmaceutics 2022, 14, 1271. https://doi.org/10.3390/pharmaceutics14061271
Kim Y, Kim H, Kim EH, Jang H, Jang Y, Chi S-G, Yang Y, Kim SH. The Potential of Cell-Penetrating Peptides for mRNA Delivery to Cancer Cells. Pharmaceutics. 2022; 14(6):1271. https://doi.org/10.3390/pharmaceutics14061271
Chicago/Turabian StyleKim, Yelee, Hyosuk Kim, Eun Hye Kim, Hochung Jang, Yeongji Jang, Sung-Gil Chi, Yoosoo Yang, and Sun Hwa Kim. 2022. "The Potential of Cell-Penetrating Peptides for mRNA Delivery to Cancer Cells" Pharmaceutics 14, no. 6: 1271. https://doi.org/10.3390/pharmaceutics14061271