You may not have a supercomputer at home, but that doesn't mean your laptop can't join the world's fastest machines in the fight against the coronavirus-caused COVID-19. A volunteer project called Folding@home has organized tens of thousands of ordinary personal computers -- like yours -- to break down big digital simulations of the virus that causes the disease into millions of bite-size chunks.
Folding@home aims to understand disease mechanisms at the molecular level then find weaknesses that medicines can exploit. Its software, for Windows, Linux and MacOS PCs, fetches a small processing job from the organization's servers, runs its calculations, then returns results to be incorporated into research studies. When it's done, your machine fetches the next job.
For two decades Folding@home has tackled diseases, including Alzheimer's and Ebola. The pandemic has bumped COVID-19 to the top of the list, though, and lots more people now want to help. The world's computers have an abundance of unused processing power. Video editors, music producers and heavy-duty gamers have to sleep, and Folding@home's distributed computing approach can keep PCs busy when their human owners aren't.
The response to COVID-19 has spurred "enormously rapid growth" during the last three weeks, with more than 700,000 new volunteers joining the 30,000 who were already active, says Greg Bowman, an assistant professor of biochemistry and molecular biophysics at Washington University in St. Louis and Folding@home's director.
Folding@home volunteers each create a short movie about the jiggling atoms that form parts of the coronavirus, each movie starting with a different arrangement of atomic motions. Having thousands of such minimovies helps Folding@home researchers then understand a virus' overall behavior.
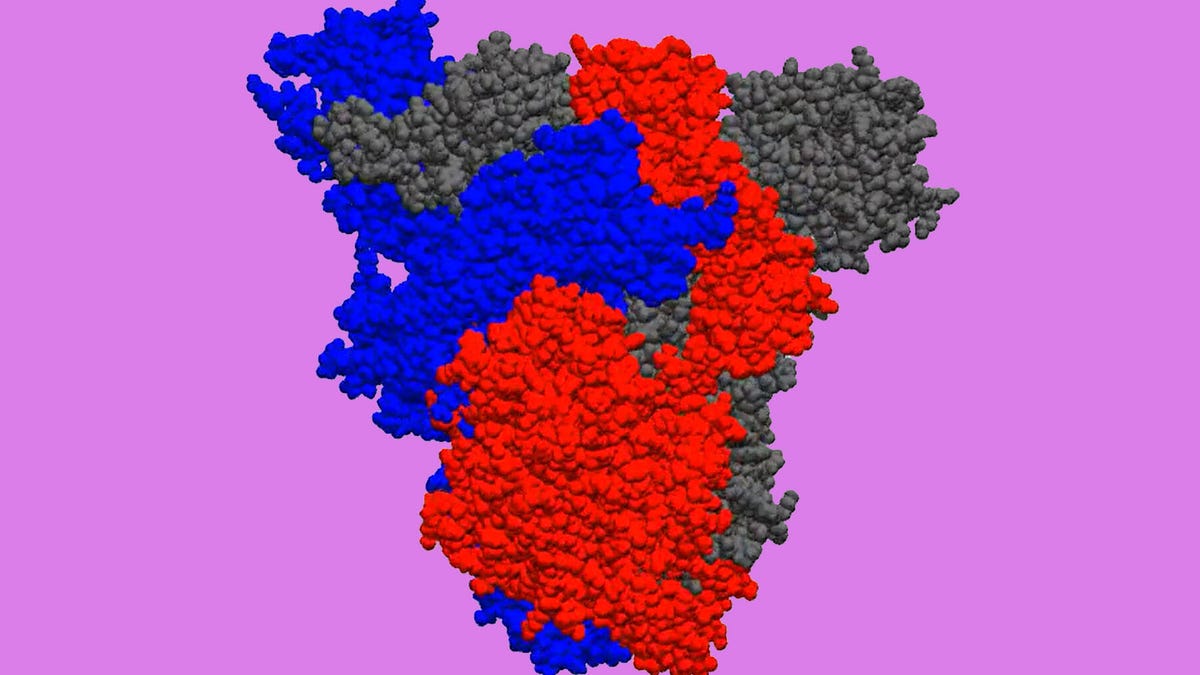
The PCs of Folding@home's volunteers create animations like this one of the new coronavirus' three-protein "spike." The animations are designed to help researchers locate pockets where drug molecules could attack.
1.5 exaflops of performance
The project can't compute as quickly as supercomputers on some important simulations. That's what enabled today's fastest machine, Summit, to recommend 77 drug compounds for experimental testing after just a few days' work. Folding@home is restricted to projects that can be broken up into mostly independent tasks.
Still, Folding@home's collective horsepower is now more than 1.5 exaflops, or a quintillion calculations per second, Bowman says. That's three quarters of the projected speed of a $600 million supercomputer called El Capitan, which is expected to be the world's fastest when it arrives in 2023.
The rush of new volunteers has overwhelmed Folding@home's servers. "The limiting thing is collecting all the data that people are sending back and writing it to disk," Bowman said. The project has doubled its server count in the last week, and more are on the way as the project woos cloud-computing partners like Oracle, Bowman said. (Oracle said it's contributing graphics-processing power to assist Folding@home research.)
Folding@home is one among many distributed-computing projects, including one called Rosetta@home that's also tackling COVID-19. Other projects are tackling astrophysics, climate prediction, particle physics, prime numbers and more.
Attacking the coronavirus spike
Viruses are built from proteins, complicated molecules assembled from genetically encoded instructions into a long string of atoms. Atomic physics determines how that string folds into a complex 3D structure. Drugs can disrupt viruses from working properly by latching onto weak points in the virus, such as pockets where a drug compound can attach.
No one can promise breakthroughs from Folding@home. And distributed computing has skeptics, such as Priya Darshan Vashishta, a University of Southern California professor and expert in atomic-level simulations, who aren't convinced the technique is an effective way to simulate the complicated reality of biochemistry.
Simulations must track the state of countless electrically interacting atoms, sometimes over relatively long distances across a protein. The protein's environment can also change its behavior.
Tracking all those interactions requires the different simulation elements to constantly communicate, and that requires a high-performance computer with fast internal communications, Vashishta says.
"This communication is very good if you have a supercomputer. That's what they're designed for," he said, adding that distributed computing can't handle the task effectively.
Finding Ebola weakness
Still, Folding@home has had successes.
One Folding@home project located a potential weakness in the Ebola virus that a drug could exploit. Another project involving antibiotic resistance located a pocket in an enzyme, a type of protein, and helped identify small drug compounds that could bind to it and inhibit its effects, Bowman said.
Folding@home is now tackling the structure of the new coronavirus, formally called SARS-CoV-2. One target is the virus' spikes, the exterior protrusions that latch onto human cells then penetrate them for infection. Each spike is made of a trio of identical proteins, Bowman said, and Folding@home's simulations are designed to model their changing configuration as their atoms wobble.
"Proteins are these little molecular machines full of moving parts," Bowman said. "We want to understand all those moving parts, find out what leads to malfunction and use that to design new therapeutics."
Folding@home is also trying to help researchers find the best drugs to attack the coronavirus by sifting through large libraries of candidates with the most promising physical properties, said John Chodera, a Folding@home participant and researcher at the Memorial Sloan Kettering Cancer Center. (He made the comments in a Reddit discussion about Folding@home on Reddit's PC Master Race forum where gamers with high-power PCs congregate.)
Making molecular movies
Even an imperfect simulation can be useful, Bowman says, likening it to the information his daughter can get out of shaking a gift-wrapped present. And the Folding@home approach is good for running many variations of the same protein under different circumstances to spot trends.
Each SARS-CoV-2 spike simulation involves 50,000 atoms surrounded by 350,000 more making up the proteins' environment.
It's nothing you could calculate on paper, but it's just what computers are good at. And what else is your laptop going to do overnight?